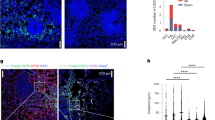
Abstract
The organization of immune cells in human tumors is not well understood. Immunogenic tumors harbor spatially localized multicellular ‘immunity hubs’ defined by expression of the T cell-attracting chemokines CXCL10/CXCL11 and abundant T cells. Here, we examined immunity hubs in human pre-immunotherapy lung cancer specimens and found an association with beneficial response to PD-1 blockade. Critically, we discovered the stem-immunity hub, a subtype of immunity hub strongly associated with favorable PD-1-blockade outcome. This hub is distinct from mature tertiary lymphoid structures and is enriched for stem-like TCF7+PD-1+CD8+ T cells, activated CCR7+LAMP3+ dendritic cells and CCL19+ fibroblasts as well as chemokines that organize these cells. Within the stem-immunity hub, we find preferential interactions between CXCL10+ macrophages and TCF7−CD8+ T cells as well as between mature regulatory dendritic cells and TCF7+CD4+ and regulatory T cells. These results provide a picture of the spatial organization of the human intratumoral immune response and its relevance to patient immunotherapy outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
MERFISH data are available at https://singlecell.broadinstitute.org/single_cell/study/SCP2510/human-lung-cancer-harbors-spatially-organized-stem-immunity-hubs-that-associate-with-response-to-immunotherapy#study-summary/. All other data are available in the article and Supplementary Information or from the corresponding authors upon reasonable request.
Code availability
Code is available at https://github.com/korsunskylab/nsclc_manuscript_2023/.
References
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734–4752 (2021).
Mihm, M. C. Jr. & Mulé, J. J. Reflections on the histopathology of tumor-infiltrating lymphocytes in melanoma and the host immune response. Cancer Immunol. Res. 3, 827–835 (2015).
Angell, H. K., Bruni, D., Barrett, J. C., Herbst, R. & Galon, J. The immunoscore: colon cancer and beyond. Clin. Cancer Res. 26, 332–339 (2020).
Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004 (2018).
Moldoveanu, D. et al. Spatially mapping the immune landscape of melanoma using imaging mass cytometry. Sci. Immunol. 7, eabi5072 (2022).
Litchfield, K. et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614 (2021).
Cristescu, R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Reschke, R. et al. Immune cell and tumor cell-derived CXCL10 is indicative of immunotherapy response in metastatic melanoma. J. Immunother. Cancer 9, e003521 (2021).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
Krishna, S. et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334 (2020).
Siddiqui, I. et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211 (2019).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1-CD8+ tumor-infiltrating T cells. Immunity 50, 181–194 (2019).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Liu, B., Zhang, Y., Wang, D., Hu, X. & Zhang, Z. Single-cell meta-analyses reveal responses of tumor-reactive CXCL13+ T cells to immune-checkpoint blockade. Nat. Cancer 3, 1123–1136 (2022).
Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021).
Oliveira, G. et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021).
Lowery, F. J. et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science 375, 877–884 (2022).
Hanada, K.-I. et al. A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers. Cancer Cell 40, 479–493 (2022).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Groom, J. R. & Luster, A. D. CXCR3 in T cell function. Exp. Cell. Res. 317, 620–631 (2011).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Wu, T. et al. The TCF1–Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Eberhardt, C. S. et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021).
Lee, J. et al. IL-15 promotes self-renewal of progenitor exhausted CD8 T cells during persistent antigenic stimulation. Front. Immunol. 14, 1117092 (2023).
Xue, D. et al. A tumor-specific pro-IL-12 activates preexisting cytotoxic T cells to control established tumors. Sci. Immunol. 7, eabi6899 (2022).
Maier, B. et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262 (2020).
Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022).
Kim, N. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020).
Song, A., Nikolcheva, T. & Krensky, A. M. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol. Rev. 177, 236–245 (2000).
Li, H. et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176, 775–789 (2019).
Dubois, S. P., Waldmann, T. A. & Müller, J. R. Survival adjustment of mature dendritic cells by IL-15. Proc. Natl Acad. Sci. USA 102, 8662–8667 (2005).
Di Pilato, M. et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530 (2021).
Cohen, M. et al. The interaction of CD4+ helper T cells with dendritic cells shapes the tumor microenvironment and immune checkpoint blockade response. Nat. Cancer 3, 303–317 (2022).
Rapp, M. et al. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J. Exp. Med. 216, 1170–1181 (2019).
Oh, D. Y. et al. Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 181, 1612–1625 (2020).
Lei, X. et al. CD4+ helper T cells endow cDC1 with cancer-impeding functions in the human tumor micro-environment. Nat. Commun. 14, 217 (2023).
House, I. G. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26, 487–504 (2020).
Chow, M. T. et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498–1512 (2019).
Nixon, B. G. et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity 55, 2044–2058 (2022).
Kersten, K. et al. Spatiotemporal co-dependency between macrophages and exhausted CD8+ T cells in cancer. Cancer Cell 40, 624–638 (2022).
Cang, Z. et al. Screening cell-cell communication in spatial transcriptomics via collective optimal transport. Nat. Methods 20, 218–228 (2023).
Cheng, H.-W. et al. CCL19-producing fibroblastic stromal cells restrain lung carcinoma growth by promoting local antitumor T-cell responses. J. Allergy Clin. Immunol. 142, 1257–1271 (2018).
Peng, Y. et al. Single-cell profiling of tumor-infiltrating TCF1/TCF7+ T cells reveals a T lymphocyte subset associated with tertiary lymphoid structures/organs and a superior prognosis in oral cancer. Oral. Oncol. 119, 105348 (2021).
Im, S. J. et al. Characteristics and anatomic location of PD-1+TCF1+ stem-like CD8 T cells in chronic viral infection and cancer. Proc. Natl Acad. Sci. USA 120, e2221985120 (2023).
Rangel-Moreno, J., Moyron-Quiroz, J. E., Hartson, L., Kusser, K. & Randall, T. D. Pulmonary expression of CXC chemokine ligand 13, CC chemokine ligand 19, and CC chemokine ligand 21 is essential for local immunity to influenza. Proc. Natl Acad. Sci. USA 104, 10577–10582 (2007).
Sato, Y. et al. Heterogeneous fibroblasts underlie age-dependent tertiary lymphoid tissues in the kidney. JCI Insight 1, e87680 (2016).
Sautès-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325 (2019).
Schumacher, T. N. & Thommen, D. S. Tertiary lymphoid structures in cancer. Science 375, eabf9419 (2022).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Wherry, E. J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007).
Hoch, T. et al. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 7, eabk1692 (2022).
Schulz, D. et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 6, 25–36 (2018).
Ardighieri, L. et al. Infiltration by CXCL10 secreting macrophages is associated with antitumor immunity and response to therapy in ovarian cancer subtypes. Front. Immunol. 12, 690201 (2021).
Alspach, E. et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701 (2019).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Ryan, N. et al. STAT1 inhibits T-cell exhaustion and myeloid derived suppressor cell accumulation to promote antitumor immune responses in head and neck squamous cell carcinoma. Int. J. Cancer 146, 1717–1729 (2020).
Meissl, K., Macho-Maschler, S., Müller, M. & Strobl, B. The good and the bad faces of STAT1 in solid tumours. Cytokine 89, 12–20 (2017).
Prokhnevska, N. et al. CD8+ T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity 56, 107–124 (2023).
Huang, Q. et al. The primordial differentiation of tumor-specific memory CD8+ T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 185, 4049–4066 (2022).
Jansen, C. S. et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019).
Grant, S. M., Lou, M., Yao, L., Germain, R. N. & Radtke, A. J. The lymph node at a glance–how spatial organization optimizes the immune response. J. Cell Sci. 133, jcs241828 (2020).
Duckworth, B. C. & Groom, J. R. Conversations that count: cellular interactions that drive T cell fate. Immunol. Rev. 300, 203–219 (2021).
Gommerman, J. L. & Browning, J. L. Lymphotoxin/light, lymphoid microenvironments and autoimmune disease. Nat. Rev. Immunol. 3, 642–655 (2003).
Piao, W. et al. Regulatory T cells condition lymphatic endothelia for enhanced transendothelial migration. Cell Rep. 30, 1052–1062 (2020).
Dejardin, E. et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17, 525–535 (2002).
Schaeuble, K. et al. Perivascular fibroblasts of the developing spleen act as LTα1β2-dependent precursors of both T and B zone organizer cells. Cell Rep. 21, 2500–2514 (2017).
Bar-Ephraïm, Y. E. & Mebius, R. E. Innate lymphoid cells in secondary lymphoid organs. Immunol. Rev. 271, 185–199 (2016).
Magen, A. et al. Intratumoral dendritic cell-CD4+ T helper cell niches enable CD8+ T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat. Med. 29, 1389–1399 (2023).
Garon, E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028 (2015).
Filbin, M. R. et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell–cell interactions. Cell Rep. Med. 2, 100287 (2021).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Korsunsky, I. et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med. 3, 481–518 (2022).
Melville, J. uwot: the uniform manifold approximation and projection (UMAP) method for dimensionality reduction. https://github.com/jlmelville/uwot (2020).
Csardi, G. et al. The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9 (2006).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. https://doi.org/10.18637/jss.v067.i01 (2015).
Gelman, A. & Su, Y. -S. arm: data analysis using regression and multilevel/hierarchical models. https://CRAN.R-project.org/package=arm (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Lee, D. T. & Schachter, B. J. Two algorithms for constructing a Delaunay triangulation. Int. J. Comput. Inf. Sci. 9, 219–242 (1980).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 (2018).
Acknowledgements
We thank the MGH Cancer Center Translational Imaging Core, MGH Pathology Department, members of the MGH spatial transcriptomics group and members of the laboratory of N.H. We thank S. X. Chao and D. Lieb for assistance with MERFISH panel design and creation of a Single Cell Portal page for the MERFISH data, respectively. This work was made possible by the generous support from Novartis as part of the MGH–Novartis Alliance. ACD donated the 12-plex RNAscope reagents as part of their Spatial Validation Program award to J.H.C. This work was also supported by public funds from the National Institutes of Health/National Cancer Institute iNCI 1T32CA207021 (to J.H.C.), 1K08CA273547-01A1 (to J.H.C.) and R00CA259511 (to K.P.), institutional funds from the MGH Fund for Medical Discovery (to J.H.C.), MGH Krantz Stewardship (to J.H.C.) and UROP funds from Massachusetts Institute of Technology (to A.L.T.) and private funds from the SU2C Phillip A. Sharp Award SU2C-AACR-PS-32 (to K.P. and N.H.), Mark Foundation for Cancer Research (to N.H.), SITC/AstraZeneca Forward Fund (to J.H.C.), Happy Lungs Foundation (to J.F.G.) and the BWH-Broad Institute Collaborative Research Award (to I.K. and N.H.). N.H. is the David P. Ryan, MD Endowed Chair in Cancer Research, a gift from Arthur, Sandra and Sarah Irving.
Author information
Authors and Affiliations
Contributions
J.H.C., L.T.N., M. Spurrell, M.M.-K., J.F.G. and N.H. conceptualized the study. J.H.C., L.T.N., V.J., J.D.P., K.Y., M.H., M.S.-F. and K.P. designed the RNA smFISH/IF and IF-only panels. J.H.C. and M. Spurrell designed the 12-plex panel. J.H.C., M. Spurrell, V.J., L.E., A.L.T. and C.L. performed multiplex and GeoMx staining and imaging. J.H.C., L.T.N., M. Spurrell, L.E., V.J. and K.H.X. performed image analysis (including cell segmentation, cell phenotyping and image co-registration). J.H.C., K.P. and N.H. designed the MERFISH panel. C.P. performed MERFISH imaging. J.H.C., L.T.N., M. Spurrell, L.E., P.M.R., R.M., S.F., A.L.T., S.S., A.M., M.P., I.Z., J.H., N.F.F., G.E., K.V.R., J.W.R., M.A. and I.K. performed computational analysis. J.H.C., C.S.N., C.B.M., J.L.B., M. Sakhi, G.M.B., M.M.-K. and J.F.G. performed clinical case finding and annotation. J.H.C., L.T.N., M. Spurrell, L.E., I.K. and N.H. wrote the manuscript. All authors edited the manuscript for intellectual content.
Corresponding authors
Ethics declarations
Competing interests
J.H.C. receives research support from the Society for Immunotherapy of Cancer AstraZeneca fellowship and a sponsored research agreement with Calico labs and previously with Novartis. J.H.C. consults for Merck and has also received a speaking honorarium from the Society for Interventional Oncology and has non-paid technology materials transfer agreements with NanoString and Vizgen. M. Spurrell owns equity in Novo Nordisk. M.P. consults for Third Rock Ventures. C.S.N. holds equity in Opko Health and receives royalties from Life Technologies and Cambridge Epigenetix. S.F.’s salary is partially supported by research funding from IBM. A.M. has served in a consultant/advisory role for Third Rock Ventures, Asher Biotherapeutics, Abata Therapeutics, ManaT Bio, Flare Therapeutics, venBio Partners, BioNTech, Rheos Medicines and Checkmate Pharmaceuticals, is currently a part-time Entrepreneur in Residence at Third Rock Ventures, is an equity holder in ManaT Bio, Asher Biotherapeutics and Abata Therapeutics, and has received research funding support from Bristol Myers Squibb. M.S.-F. receives funding from Bristol Myers Squibb. G.M.B. has sponsored institutional research agreements with Olink Proteomics, Teiko Bio, InterVenn Biosciences and Palleon Pharmaceuticals. G.M.B. served on advisory boards for Iovance, Merck, Nektar Therapeutics, Novartis and Ankyra Therapeutics. G.M.B. consults for Merck, InterVenn Biosciences, Iovance and Ankyra Therapeutics. G.M.B. holds equity in Ankyra Therapeutics. M.J.A has financial and consulting interests unrelated to this work in SeQure Dx and Chroma Medicine. M.M.-K. has served as a consultant for AstraZeneca, BMS, Sanofi and Janssen Oncology and receives royalties from Elsevier. J.F.G. has consulted and/or had advisory roles for Agios, Amgen, AstraZeneca, Array BioPharma, Blueprint Medicines Corporation, BMS, Genentech/Roche, Gilead Sciences, Jounce Therapeutics, Lilly, Loxo Oncology, Merck, Mirati, Silverback Therapeutics, Sanofi, GlydeBio, Curie Therapeutics, Novartis, Moderna Therapeutics, Oncorus, Regeneron, Takeda and Pfizer; has stock and ownership in Ironwood Pharmaceuticals; and has received Honoraria from Merck, Novartis, Pfizer and Takeda; and institutional research funding from Adaptimmune, ALX Oncology, Merck, Array BioPharma, AstraZeneca, Blueprint Medicines Corporation, BMS, Scholar Rock, Genentech, Jounce Therapeutics, Merck and Novartis; research funding from Novartis, Genentech/Roche and Takeda, and has an immediate family member who is an employee of Ironwood Pharmaceuticals. N.H. holds equity in and advises Danger Bio/Related Sciences, is on the scientific advisory board of Repertoire Immune Medicines and CytoReason, owns equity and has licensed patents to BioNtech, and receives research funding from Bristol Myers Squibb and Calico Life Sciences. J.H., N.F.F. and G.E. are employees of Vizgen. C.P. (now at Takeda) was an employee of Vizgen when this research was conducted. J.W.R. and K.V.R. are employees of NanoString. I.K. receives research funding from the Chan Zuckerberg Initiative and has consulting/advisory roles with Mestag Therapeutics and Scailtye. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Sarah Teichmann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: N. Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Leiden clustering of immunity hubs.
(a–d) Paired RNA smFISH/IF and IF only stained slides were coregistered for 46 of the patients. PanCK staining is shown in cyan for RNA smFISH/IF and magenta for IF panel images. (e–g) tSNE projection as in Fig. 2a colored by (E) patient ID, (F) patient response status, and (G) immunity hub size. (h) Patient composition within each immunity hub Leiden subcluster. (i) Plot as in (H), but the y-axis depicts the number of subclusters, colored by the patient. Each column is a Leiden subcluster. (j) Frequency of immunity hub subclusters expressed as a fraction of total tumor area for responders and non-responders (NR = 33 and R = 13 patients). Two-sided Mann-Whitney test p-value is shown. (k-l) Kaplan-Meier analysis of progression-free survival (PFS) and overall survival (OS) for each subcluster. Patients were classified as either above or below a threshold of 0.1% of total tumor area for each subcluster. Shown p-values are adjusted for multiple hypothesis testing (7 tests) using the Benjamini-Hochberg method (J-L). (m) Counts of indicated phenotypes within immunity hubs, as in Fig. 2g. Phenotype counts were normalized by immunity hub area and scaled from 0-1. Each point represents the mean value across all immunity hubs of a given Leiden subcluster for each patient having that subcluster (number of patients having subcluster: #1 = 18, #2 = 33, #3 = 31, #4 = 11, #5 = 27, #6 = 25, and #7 = 30). Boxplots are defined as follows: center line = median, box minima and maxima = interquartile range (IRQ), and whiskers = 1.5*IQR. Statistical comparison was performed using an unpaired two-sided Mann-Whitney test relative to subcluster 3. Benjamini-Hochberg adjusted p-values shown: ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (n) Fraction of TCF7+PD1+ CD8 T cells found within subcluster 3 in responders versus non-responders. Each dot represents one patient (NR = 33 and R = 13 patients). Two-sided Mann-Whitney test p-value is shown. (o) Counts of contact (at least touching in a cardinal direction) between TCF7+ aggregates and subcluster 3 immunity hubs versus other immunity hub subclusters (subclusters 1-2 and 4–7). Statistical comparison was performed using a two-sided Fisher’s exact test.
Extended Data Fig. 2 Representative images for each immunity hub subcluster.
Each row features an immunity hub from a different patient among the 46 patients with successfully coregistered panels. Aligned serial sections are shown for RNA ISH/IF panel (Column A), H&E (Column B), and IF only panel (Columns C-G). (a) IFNG+ cells (circles) are observed in immunity hubs. (b) H&E shows immunity hubs contain immune infiltrate abutting neoplastic cells. (c) Ki67+CD8+ (circles) and Ki67+TCF7+CD8+ cells (arrows) are found. (d) Ki67+TCF7+CD8+ (arrows), TCF7+CD8+ (circles), and TCF7+PD1+CD8+ cells (arrowheads) are found. (e) PD1+CD8+ (circles) and TCF7+PD1+CD8+ cells (arrowheads) are found. (f) Composite image showing indicated channels. (g) PD-L1+ cells are found.
Extended Data Fig. 3 Cell-based clustering identifies many immune cell types in MERFISH data.
(a) Histogram of transcripts per cell by patient. (b) UMAP of MERFISH data from Fig. 3b, colored by patient. (c) Cell counts by coarse cell type for each patient. (d) UMAP of immune cell MERFISH data from Fig. 3b, colored by patient. (e) Cell counts for fine immune clustering for each patient. (f) Heatmap including both coarse and fine cell clusters versus expression of key marker genes.
Extended Data Fig. 4 Cell-based analysis of MERFISH data reveals cell populations enriched within the stem-immunity hubs.
(a) The tile grid consisted of regular hexagons with area = 2500 μm2. To improve smoothness in tile clustering, hexagonal tiles were dilated with a 15 μm buffer. (b) Transcripts per non-dilated 2500 μm2 tile by patient. (c) Spatial map of tumor for four MERFISH samples, with tiles colored by four region types. (d) Mean number of cells per tile per aggregate for indicated clusters. Each dot represents one patient, colored by patient number. The open gray circle represents the mean cell per tile count for all aggregates of the indicated tile type across patients. (e) Forest plots depict the log2 odds ratio of the abundance (normalized to physical area) of a cell type in the indicated region type versus abundance in library-matched tumor areas. Error bars denote the 95% confidence interval (n = 4 patients, as in (D)) and center dot denotes point estimate.
Extended Data Fig. 5 Co-localization of cells.
(a) Neighboring cell analysis for cells within stem-immunity hubs, as in Fig. 5. For each plot, indicated cluster position (row) is held constant and enrichment with partner (column) is quantified versus scrambled distribution. Mean z-scores across all four samples are shown. Two-tailed p-values were calculated from the mean z-scores and FDRs computed with the Benjamini-Hochberg procedure across all p-values. FDR < 0.05 are denoted with an asterisk. Total number of indicated index cells is shown at right, based on Supplementary Table 1,h. (b) Absolute frequency of interactions (mean of the mean frequencies across the four samples) for a given index cell (row) and partner cell type (column). (c) The red line on histograms shows how frequently specific T cell types are observed to neighbor the indicated myeloid cell type within the stem-immunity hubs for indicated patients. Gray bars denote random distribution (expected distribution based on cell type frequency).
Extended Data Fig. 6 Gene signatures of T cells neighboring mreg DCs and CXCL10+ macrophages.
Heatmap of gene expression of CD4 T cells, CD8 T cells, and Tregs, subdivided into three groups reflecting their neighborhood composition: adjacent to at least one LAMP3+CCL19+ mreg DC, adjacent to at least one CXCL10+ macrophage, or adjacent to neither. T cells adjacent to both were rare and thus not included in this analysis. Only genes significantly differentially overexpressed (GLMM adj. p < 0.05 & logFC > 0.5, see Methods) in one of the rows are shown.
Extended Data Fig. 7 Cell-cell interactions within stem-immunity hubs.
Neighboring cell analysis for cells within stem-immunity hubs based on 12-plex RNAscope data. For each plot, indicated cluster position (row) is held constant and enrichment with partner (column) is quantified versus scrambled distribution. Z-scores were calculated from the mean and standard deviation of the permutation-based empirical null distribution. Mean z-scores across all four samples are shown. Two-tailed p-values were calculated from the mean z-scores and FDRs computed with the Benjamini-Hochberg procedure across all p-values. FDR < 0.05 are denoted with an asterisk.
Extended Data Fig. 8 TCF7+ regions feature CCL19, CCL22, and variable MS4A1, and frequently overlap with immunity hub subcluster 3.
(a–c) Six cases (R = 4, NR = 2) from the 68 patient cohort with TCF7+ aggregates (smFISH/IF panel staining shown in B) were stained by H&E (C) and an additional panel for CCL19, CCL22, MS4A1, CXCL10/11, and PanCK. Dashed ovals indicate areas with CXCL10/11+ cells. Arrows point to CCL22+ cells.
Extended Data Fig. 9 Representative NanoString GeoMx regions of interest.
(a) Fluorescently labeled RNA smFISH probes for CXCL10 and CCL19 on GeoMx slides guided region of interest (ROI) selection for transcriptional profiling. Histological features, region location, and number of regions profiled are noted. (b) Representative GeoMx images of stem-immunity (SI) hub, non-stem-immunity hub, and TLS ROI (as defined in (A)) for all five patients profiled using Nanostring GeoMx. White lines indicate ROI boundaries. ROI ID number for a given patient is indicated in the top left corner of each image.
Extended Data Fig. 10 Transcriptional profile of stem-immunity hubs, non-stem-immunity hubs, and TLS using GeoMx.
(a, b) Comparison of (A) stem-immunity hub and non-stem-immunity hub and (B) non-stem-immunity hub and TLS gene expression using GeoMx CTA assay. Volcano plots depict pooled ROIs across all 5 samples. Each dot represents one gene. Dot coloring is as follows: blue - ISGs, green - myeloid genes, purple - T cell genes, and orange - B cell genes. (c) Scatter plot of expression for 177 genes included in both GeoMx and MERFISH panels for manually annotated stem-immunity hubs (GeoMx) and tile cluster-defined stem-immunity hubs (t7, MERFISH) (pearson correlation r = 0.78, p < 1E-35). Axes indicate log2 fold-change of gene expression in stem-immunity versus non-stem-immunity hub. Each dot denotes one gene. Red dots denote the top 10 over-expressed genes in stem-immunity (versus non-stem-immunity hub) in the GeoMx dataset.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J.H., Nieman, L.T., Spurrell, M. et al. Human lung cancer harbors spatially organized stem-immunity hubs associated with response to immunotherapy. Nat Immunol 25, 644–658 (2024). https://doi.org/10.1038/s41590-024-01792-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-024-01792-2
This article is cited by
-
Spatiotemporal metabolomic approaches to the cancer-immunity panorama: a methodological perspective
Molecular Cancer (2024)
-
Spatial biology captures the effects of neoadjuvant chemo-immunotherapy in lung cancer
Nature Genetics (2024)
-
T cell dysfunction and therapeutic intervention in cancer
Nature Immunology (2024)
-
Immunity hubs orchestrating antiviral defense
Cell Research (2024)
-
Tertiary lymphoid structures in anticancer immunity
Nature Reviews Cancer (2024)